Chapter 15 Model selection
15.1 Introduction
If there is only one, or a small number of possible explanatory variables (also known as covariates, predictors or independent variables) choosing a linear regression model can be straightforward. What happens when there are many explanatory variables? Some covariates may be more important/useful than others in explaining the response variable. Since conclusions depend, to some extent, on the covariates in the linear model, how do we decide which covariates to include? This chapter considers how to choose between competing models using different model selection procedures: \(p\)-values, fit scores and automated methods. The concepts described in this chapter use examples introduced in Chapter 14.
15.1.1 Criteria for model selection
We want to have an appropriate set of covariates in our model:
- if we include too few variables we throw away valuable information, and
- if we include non-essential variables the standard errors and \(p\)-values tend to be too large.
- if the models are too simple (under-fitted), or too complex (over-fitted), the models will have poor predictive abilities.
We want to include variables which:
- have a genuine relationship with the response and
- offer a sufficient amount of new information about the response (given the variables already included)
We want to exclude variables that:
- offer essentially the same information about the response; e.g., we want to avoid collinearity.
15.2 Collinearity
Collinearity is a linear association between two variables in a linear regression model. ‘Multicollinearity’ refers to linear associations between two or more variables in a multiple regression model.
When ‘collinear’ variables are fitted together in a model,
- the resulting model is unstable (because we are trying to estimate two parameters when one will do) and,
- we obtain inflated standard errors for these estimates.
We have methods to detect this however.
15.2.1 Variance inflation factors
Collinearity can be detected using ‘variance inflation factors’ (VIFs). These are based on fitting linear models between each covariate (in turn) and the remaining covariates and assessing the predictive power of each:
\[\begin{equation} VIF_p=\frac{1}{1-R_p^2} \end{equation}\]
where \(R^2_p\) is the squared correlation between the \(p\)-th observed covariate value and those predicted by a linear model containing the other covariates. If any of the \(R^2_p\) values are high, then the VIF will also be high.
There are no firm rules about how large VIFs need to be before remedial action (e.g. removing a covariate) is required; some say VIFs \(> 5\), some say VIFs \(>10\).
VIFs require adjustment if we estimate multiple parameters (i.e. regression coefficients) for a particular covariate, for example, the number of regression coefficients estimated for a factor is the ‘number of levels - 1.’ However, VIFs are easily calculated using software.
15.2.1.1 Doing this in R
We consider a regression of total height on left leg length (LLL) and right leg length (RLL) (seen in Chapter 14). The covariates LLL and RLL are strongly correlated but the variable length of hair (LOH) is not correlated with left leg length (see below) or indeed total height.
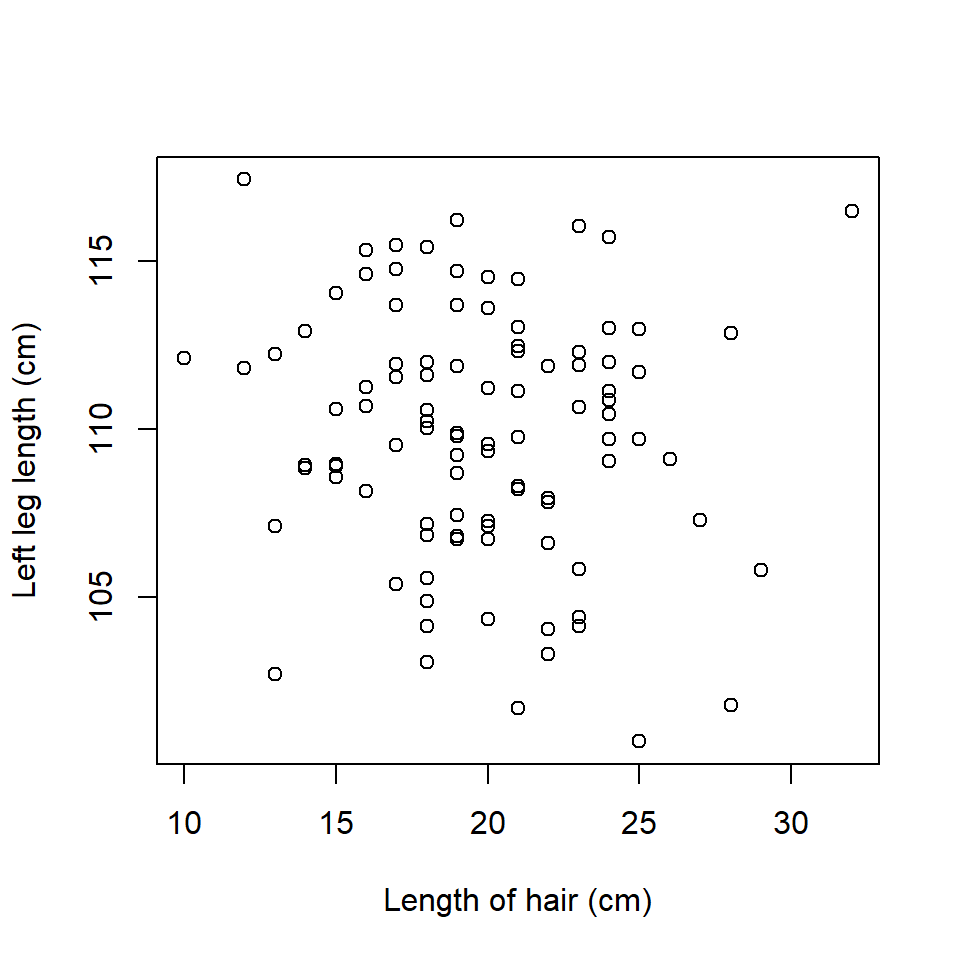
Figure 15.1: Relationship between length of hair and left leg length.
A function to obtain VIFs is available in the car package. To calculate the VIFs, we need to first fit a linear model with all the potential explanatory variables included.
# Fit multiple regression model
modelAll <- lm(TotalHeight ~ LLL + RLL + LOH)
# Load package
require(car)
# Calculate VIFs
vif(modelAll) LLL RLL LOH
5636.487167 5637.364338 1.014372 The large values of the VIFs for LLL and RLL indicate that they are (not surprisingly) highly correlated. This suggests removal of one of the leg length variables. The variable RLL is removed from the model and the process is repeated. The resulting VIFs indicate that the remaining variables are not collinear.
# Fit multiple regression model
model2vars <- lm(TotalHeight ~ LLL + LOH)
# Calculate VIFs
vif(model2vars) LLL LOH
1.011245 1.011245 NOTE: Variables should be removed one at a time and everything retested.
15.2.1.2 Dealing with collinearity
Collinearity can be addressed by removing one of the collinear variables, but alternative methods exist if it desirable to retain the full set of covariates.
The removal of one, or more, collinear covariates may occur automatically if \(p\)-values (see below) are used to drop terms from a model. This occurs since collinear terms are often unstable and thus highly uncertain, which means the associated \(p\)-values are often large. A 5 minute clip about this issue can be found here. A large \(p\)-value could mean either the variable has no effect, or is correlated with another predictor.
Alternatively, the analyst may use their judgement in which collinear covariate(s) are retained and which are omitted (but readers might be skeptical of such a subjective approach). This can be done, for instance, by comparing the relative predictive power of the model with and without each covariate and choosing the covariate which predicts the response `best’.
Q15.1 Does a low VIF indicate that a variable should be in the final model?
15.3 \(p\)-value based model selection: the \(F\) test
Collinearity identifies correlated predictor variables, it does not necessarily generate a good model. The \(p\)-value associated with an estimated regression coefficient can be used to decide whether to include a particular variable in the final model. Essentially, we perform a hypothesis test where the null hypothesis is that the regression coefficient is equal to zero; a regression coefficient equal to zero would have the effect of eliminating that variable from the model. The \(p\)-value associated with a relevant test statistic is then interpreted in the usual way.
For covariates with one associated coefficient, retention can be based on the value of the associated \(p\)-value (i.e. large \(p\)-values suggest omission, small \(p\)-values (e.g. \(<\) 0.05) suggest retention).
For variables with multiple coefficients (e.g. factors) we are interested in assessing a group of coefficients simultaneously. In chapter 14, a model was fitted which included
monthas a factor variable; it had four levels and so there were three regression coefficients associated with it - these were denoted in the model as \(\beta_5\), \(\beta_6\), \(\beta_7\). The test of interest is:
\[\begin{align*} H_0:~& \beta_5=\beta_{6}=\beta_{7}=0\\ H_1:~& \textrm{ at least one of } \beta_5,\beta_{6},\beta_{7} \neq 0 \end{align*}\]
We look at an example of this now.
15.3.0.1 Comparing models with and without a factor
In fitting a linear model to the EIA data, Month was treated as a factor variable with four levels, therefore, models with and without Month differ by 3 parameters. We wish to compare a reduced model (without month) with the full model (with month) using a significance test. The idea is as follows:
- If month is an important predictor then a model with month should predict the response values considerably better than a model without month.
- If a model with and without month are equivalent then models with and without month will predict the response data similarly.
We can use an \(F\) test to formally test the hypothesis that the model without month (with \(q\) parameters) is as good as the model with month (with \(P\) parameters) and hence the smaller model is preferred (and month is not required).
Analysis of Variance Table
Response: Density
Df Sum Sq Mean Sq F value Pr(>F)
XPos 1 82435 82435 106.3265 < 2.2e-16 ***
YPos 1 3545 3545 4.5719 0.03251 *
DistCoast 1 129640 129640 167.2118 < 2.2e-16 ***
Depth 1 95065 95065 122.6164 < 2.2e-16 ***
Phase 2 5956 2978 3.8413 0.02148 *
as.factor(Month) 3 49779 16593 21.4019 7.656e-14 ***
Residuals 31492 24415800 775
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Note that the explained sum of squares produced by the anova command is sequential so if we want to test for Month given all the other terms in the model i.e Month must come last in the list of variables (as it is in this example).
Differences in average density across months
What can we conclude from these results?
- This test provides a large \(F\) test statistic (21.2102) and small associated \(p\)-value (\(p<0.0001\)) for
Month, which suggests a model withMonthis significantly better than a model withoutMonth. - This indicates genuine month to month differences in average density and thus month information should be retained in the model.
Differences in average density across phases
Phase of construction was included as another factor variable in the model; this had three levels (denoted by A, B and C). If we re-order the model so Phase is last, we can consider whether Phase should be in the model.
Analysis of Variance Table
Response: Density
Df Sum Sq Mean Sq F value Pr(>F)
XPos 1 82435 82435 106.3265 < 2.2e-16 ***
YPos 1 3545 3545 4.5719 0.032509 *
DistCoast 1 129640 129640 167.2118 < 2.2e-16 ***
Depth 1 95065 95065 122.6164 < 2.2e-16 ***
as.factor(Month) 3 46310 15437 19.9105 6.894e-13 ***
Phase 2 9425 4713 6.0784 0.002294 **
Residuals 31492 24415800 775
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1For phase, the \(F\) test results are quite the opposite to that for
Month: \(F=0.2409\) and \(p=0.7859\). This indicates no significant difference between models with and withoutPhaseand, therefore, no genuine differences in average density across phases.This suggests phase should be omitted from the model given the presence of the other variables.
In the above examples, we have written the model with the covariates in different orders to ascertain the appropriate \(p\)-values. The R function Anova (as opposed to anova) gives the \(p\)-values for all covariates assuming the term is the last in the model. Thus:
Anova Table (Type II tests)
Response: Density
Sum Sq Df F value Pr(>F)
XPos 74801 1 96.4802 < 2.2e-16 ***
YPos 121103 1 156.2012 < 2.2e-16 ***
DistCoast 16028 1 20.6738 5.466e-06 ***
Depth 96320 1 124.2357 < 2.2e-16 ***
Phase 9425 2 6.0784 0.002294 **
as.factor(Month) 49779 3 21.4019 7.656e-14 ***
Residuals 24415800 31492
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The \(p\)-values are now equivalent to having the relevant term as the last term in the model, even though Depth is the actual last term now. Note the brackets with the words Type II tests - this indicates the so called ‘Type II’ sum of square’s are being used (sums of squares that assume each term is the last in the model) as opposed to ‘Type I.’ Type I errors use the sequential sum of squares (which is affected by the order of the variables in the model specification) and are provided by the function anova.
15.4 Relative model fit
While \(F\) tests (and associated \(p\)-values) can be used to compare nested models (when one model is a special case of the other) they cannot be used to compare models which are not nested. In contrast, both nested and non-nested models can be compared using `information-based’ fit criteria such as the AIC or BIC statistic.
15.4.1 Akaike’s Information Criterion (AIC)
Occam’s Razor is the very useful rule that when comparing models of equal explanatory power (i.e. models have the same \(R^2\)), that one should choose the simplest (i.e. fewest parameters). But what if the models have different levels of complexity along with different levels of explanatory power? Is a simple model of low explanatory power better than a complicated model with high explanatory power?
The AIC statistic is a fit measure which is penalized for the number of parameters estimated in a model;
- a smaller AIC value signals a better model.
The AIC is calculated using:
\[\textrm{AIC} = \textrm{Fit to the data} + \textrm{model complexity}\]
\[\begin{equation}\label{eq:aic} AIC= -2 \textrm{ log-likelihood value} + 2P \end{equation}\] Where
- ‘Fit to the data’ is measured using the so-called `log-likelihood’ value calculated using the estimated parameters in the model,
- ‘model complexity’ is measured by \(2P\) where \(P\) is the number of covariates used to fit the model.
15.4.1.1 AICc
When the sample size is not a great deal larger than the number of parameters in the model, the small sample corrected AICc is a better measure than AIC:
\[\begin{equation}\label{eq:aicc} AICc=AIC+\frac{2P(P+1)}{n-P-1} \end{equation}\]
This value gets very close to the AIC score when sample size, \(n\), is much larger than \(P\).
15.4.1.2 BIC
The BIC score differs from the AIC score by employing a penalty that changes with the sample size (\(n\)):
\[\begin{equation} BIC=-2 \textrm{log-likelihood value} + \log(n)P \end{equation}\]
As for the AIC and AICc, smaller values signal `better’ models. BIC is more conservative than AIC and will produce models with fewer variables.
15.5 Other methods of model selection
Information criterion and \(p\)-values represent two approaches to model selection both based on likelihood. However there are other methods. Cross-validation is a method used both by statisticians and data scientists, where a model is fitted to a subset (the “training set”) of the available data and then tested (“validated”) against the remainder of the dataset, the “validation set.” There a variety of forms of cross-validation.
15.6 Automated variable selection
The number of possible combinations soon increases as the number of explanatory variables increases. There are various procedures described below which can be used to select the ‘best’ model. All of the procedures could be implemented manually (i.e. by fitting a model, obtaining a test statistic, fitting the next model, etc.) but this can be very time-consuming. Fortunately, there are R functions available which can be used to implement them.
15.6.1 Stepwise selection
Stepwise selection is a commonly used automated method which adds and drops covariates (from some start point) one at a time until no change occurs in the selected model.
- ‘Importance’ of variables can be measured in a number of ways; the AIC/AICc and BIC statistics are commonly used, alternatively \(p\)-values could be used.
Selection proceeds either:
- forwards from a simple model by addition of covariates, or
- backwards from a complex model by dropping covariates.
- More elaborate algorithms are possible.
In forward selection, one variable is added at a time and the AIC etc. is calculated. Then another variable is tried INSTEAD, until all the candidate variables have been tried. The model with the lowest AIC (if lower than the starting model) is selected. A new ‘round’ of selection then begins with the remaining candidate models considered. Modelling proceeds until no further reduction in AIC is found.
In backward selection, starting from a model with all potential variables, one variable is removed and the AIC etc. is calculated. Then another variable is removed INSTEAD, until all the candidate variables have been tried. The model with the lowest AIC (if lower than the existing start model) is selected. A new ‘round’ then begins with the remaining candidate variables removed one at time then replaced as before. Modelling proceeds until no further reduction in AIC is found.
These two methods will not necessarily select the same model because different combinations of variables are being included in the considered models.
15.6.2 All possible subsets selection
Rather than rely on an algorithm to determine the order in which variables are picked and dropped from a model (which can affect which covariates are retained), we can compare ‘fit scores’ for all possible models.
For example, for a 4 covariate model we can compare the fit scores for:
- an intercept only model (1 model)
- all models containing one covariate (4 models)
- all models containing two covariates (6 models)
- all models containing three covariates (4 models)
- the full model with 4 covariates (1 model)
However, this method becomes prohibitively time consuming when there are a lot of covariates.
15.6.2.1 Doing this in R
One way to fit all possible models is to use the dredge function inside the MuMIn library. The default score for ranking models is the AICc (but this can easily be changed) and the output includes all possible models which are ordered by AICc.
As an example, we return to a model fitted to the EIA data with six potential explanatory variables. The dredge function has been used; the R code is shown (but not executed because it generates a lot of output). The regression coefficients for models with the four lowest AICc scores are shown below. :
require(MuMIn)options(na.action = "na.fail")
dredge(modelEIAall)Fixed term is "(Intercept)"
Global model call: lm(formula = Density ~ XPos + YPos + DistCoast + Depth + Phase +
as.factor(Month) + Depth, data = wfdata)
---
Model selection table
(Int) as.fct(Mnt) Dpt DsC Phs XPs YPs df logLik AICc delta weight
64 3286 + -0.4544 -0.3152 + 0.1175 -0.5549 11 -149489.8 299001.6 0.00 0.983
56 3288 + -0.4532 -0.3213 0.1185 -0.5553 9 -149495.9 299009.7 8.16 0.017
60 2702 + -0.5575 + 0.1009 -0.4567 10 -149500.1 299020.2 18.67 0.000
52 2692 + -0.5583 0.1016 -0.4552 8 -149506.6 299029.2 27.65 0.000
63 3260 -0.4524 -0.3117 + 0.1184 -0.5505 8 -149521.9 299059.7 58.16 0.000
55 3264 -0.4513 -0.3174 0.1193 -0.5513 6 -149525.7 299063.4 61.82 0.000
Models ranked by AICc(x)We observe:
- the best model (by AICc) is at the top,
- estimated regression coefficients are displayed for continuous variables and a
+for factor variables; neither a coefficient or a+indicates the variable is excluded. For example, line 3 showsDistCoastomitted and line 5 showsMonthomitted. deltais the difference in AICc between the best (top) model and the listed model,weightassociated with fit score (described below).
One thing to note is that records with any missing values need to be excluded before using dredge because models with different numbers of observations cannot be compared with information criteria..
15.6.2.2 AIC weights
It is always important to be sensible about the covariates considered for selection, but this method also allows model comparison using weights based on your chosen fit score (e.g. AIC/AICc/BIC).
These weights are based on the relative size of the difference between the fit of each candidate model and the best model using your chosen fit score. For example, using the AIC, weights are given by:
\[\begin{equation}\label{AICWeights} w_i(AIC)=\frac{\exp\{-\frac{1}{2}\Delta_i(AIC)\}}{\sum_{k=1}^K\exp\{-\frac{1}{2}\Delta_k(AIC)\}} \end{equation}\]
where \(K\) is the number of models considered and \[ \Delta_i(AIC)=AIC_{i}- \textrm{minimum AIC} \]
These weights sum to one over all candidate models and can be calculated using your chosen fit score e.g. AIC, AICC or BIC statistics.
15.7 Example: model selection with the medical data
We will now consider model selection in the medical data set. This data set has an interesting diversity of variable types.
As a reminder, let’s look a the data available in the TEON data set:
head(meddata) gend age vitdresul vitdc vit.12 vitbc folate TEON teonpres
1 female 50 10.98 insufficiency 310.0 normal 19.17 Yes 1
2 female 39 13.46 insufficiency 238.0 normal 8.16 Yes 1
3 female 39 15.36 insufficiency 361.0 normal 5.55 Yes 1
4 male 28 11.32 low 113.4 low 4.58 Yes 1
5 male 17 5.88 defficiency 313.0 normal 3.18 Yes 1
7 male 26 12.21 insufficiency 986.0 high 16.41 Yes 1Let’s consider creating a linear model with some covariates of mixed types. Specifically, we’ll look at folate , TEON (presence/absence of TEON), and gend (gender) to explain vitdresul (vitamin D level). The data is shown in Figure 15.2.
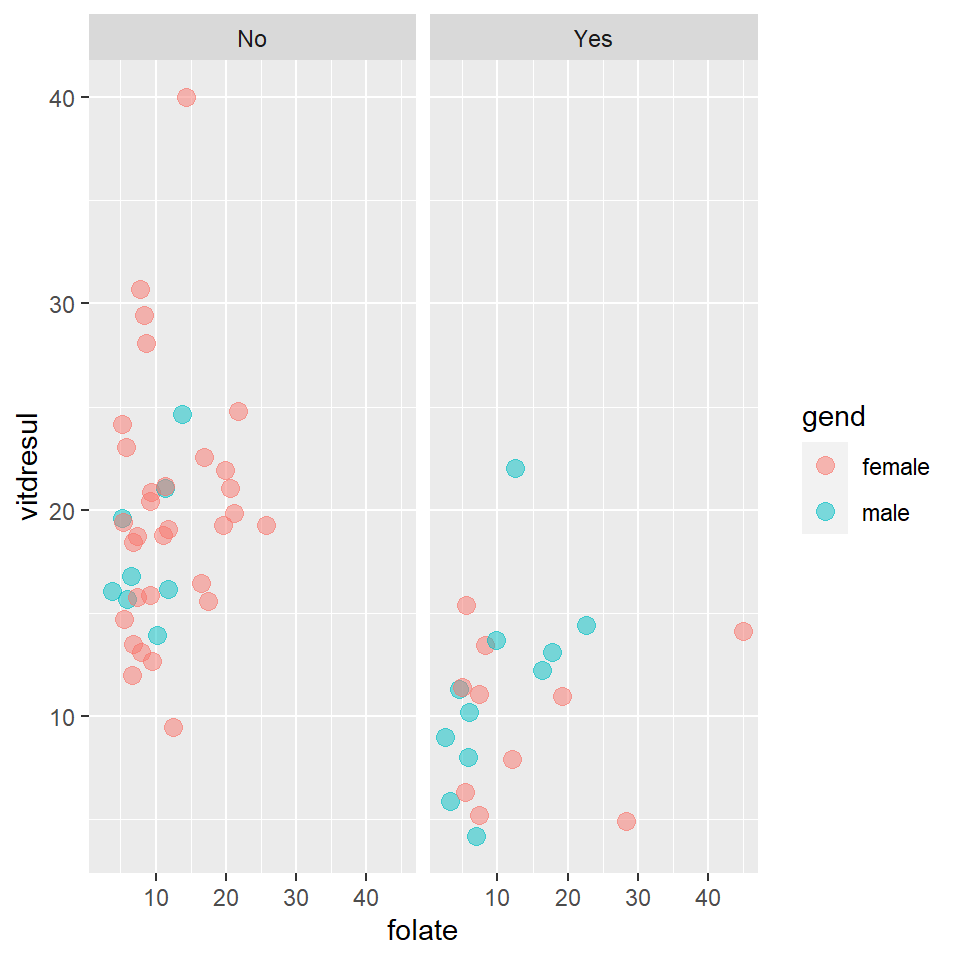
Figure 15.2: Scatterplot showing the relationships between vitamin D level, folate, TEON and gender.
15.7.1 Model specification
Fitting models with lm is straight-forward. We specify response, \(y\), as a function of several explanatory variables, \(x\) - here vitdresul is a function of gend, TEON, (both categorical) and folate (numeric). Note, we may have causality the wrong way around here as TEON may be a consequence of vitamin deficiency (but this model serves to illustrate the methods).
multiReg_lm <- lm(vitdresul ~ TEON + gend + folate, data=meddata)We request a summary, as for other lm
summary(multiReg_lm)
Call:
lm(formula = vitdresul ~ TEON + gend + folate, data = meddata)
Residuals:
Min 1Q Median 3Q Max
-10.3112 -3.4375 -0.5539 1.6053 20.0017
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 18.27764 1.46004 12.519 < 2e-16 ***
TEONYes -8.95244 1.53647 -5.827 2.91e-07 ***
gendmale -0.10386 1.60546 -0.065 0.949
folate 0.11901 0.09539 1.248 0.217
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.322 on 56 degrees of freedom
Multiple R-squared: 0.4153, Adjusted R-squared: 0.3839
F-statistic: 13.26 on 3 and 56 DF, p-value: 1.192e-0615.7.2 Interpreting the parameter estimates
As we have factor covariates, we have to interpret the model coefficients with respect to some baseline level(s). Note TEON = no and Gender = Female are not listed in the estimates - this is the baseline (Table 15.1).
| TEON | Gender | Fitted model |
|---|---|---|
| No | Female | \(\widehat{\textrm{vitdresul}} = 18.278 + 0.119\textrm{folate}\) |
| Yes | Female | \(\widehat{\textrm{vitdresul}} = 18.278 -8.952 + 0.119\textrm{folate}\) |
| No | Male | \(\widehat{\textrm{vitdresul}} = 18.278 -0.104 + 0.119\textrm{folate}\) |
| Yes | Male | \(\widehat{\textrm{vitdresul}} = 18.278 -8.952 -0.104 + 0.119\textrm{folate}\) |
The intercept is 18.27:
this is the estimated mean of
vitdresulwhenTEON= nogender= female andfolate= 0
Further, this is significantly different from zero (\(p\)-value is \(<2\times 10^{-16}\) - effectively zero).
The TEONYes parameter is -8.95:
- this is the difference from the intercept, moving from
TEON= No toTEON= yes - it is a decrease of 8.95 units which is statistically significant (\(p\)-value = \(2\times 10^{-7}\)).
The gendmale parameter is -0.104:
- this is the difference from the intercept, moving from
gender= Female togender= Male - it is a decrease of 0.104 units but is not statistically significant (\(p\)-value = 0.949).
The coefficient for folate is 0.119:
- this is the mean increase in
vitdresulfor a unit increase infolate - the increase is 0.119 units; not statistically significant (\(p\)-value = 0.217).
It is easy to get 95% CIs for the parameter estimates:
confint(multiReg_lm) 2.5 % 97.5 %
(Intercept) 15.35283815 21.2024493
TEONYes -12.03035966 -5.8745290
gendmale -3.31998297 3.1122594
folate -0.07208286 0.3101002Consistent with the tests previously (because we’re using 0.05 as the p-value cutoff):
- zero is not a plausible value for the intercept, or for the
TEONrelationship - zero is a plausible value for the
folateandgendereffects.
Possibly we could remove some variables from the model.
15.7.3 What ‘should’ be in the model?
We’ll now look to select components from/for our model using the methods described previously:
- Selection by p-values
- Selection by AIC and similar measures
- Automated selection - forwards, backwards and all-possible-subsets
First, let’s try including all variables that are available in the medical data set to explain vitdresul. The . in the model formula is shorthand for that.
# First fit a model with mixed types and reiterate the interpretation component
library(car)
# Fit everything
bigReg <- lm(vitdresul ~ ., data=meddata)summary(bigReg)
Call:
lm(formula = vitdresul ~ ., data = meddata)
Residuals:
Min 1Q Median 3Q Max
-6.604 -2.603 0.000 1.869 10.845
Coefficients: (1 not defined because of singularities)
Estimate Std. Error t value Pr(>|t|)
(Intercept) 8.415038 7.653793 1.099 0.276940
gendmale 0.561497 1.269939 0.442 0.660330
age -0.011608 0.042999 -0.270 0.788315
vitdcinsufficiency 7.186854 1.814761 3.960 0.000242 ***
vitdclow 6.451302 4.585442 1.407 0.165766
vitdcnormal 23.705316 3.405476 6.961 7.63e-09 ***
vit.12 0.000514 0.005760 0.089 0.929249
vitbclow 1.036309 8.309493 0.125 0.901260
vitbcnormal 2.811365 5.600409 0.502 0.617921
folate 0.060777 0.078379 0.775 0.441811
TEONYes -5.155763 1.494984 -3.449 0.001167 **
teonpres NA NA NA NA
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.963 on 49 degrees of freedom
Multiple R-squared: 0.7162, Adjusted R-squared: 0.6583
F-statistic: 12.37 on 10 and 49 DF, p-value: 2.451e-10In the summary output above, not all regression coefficients have been estimated (specified by NA) and there is a message about ‘singularities.’ What has happened?
- Specifying
y ~ .should put everything in the data set as a covariate (except the response, obviously). - The reason is that we effectively have two (same) covariates indicating the presence/absence of TEON -
TEONandteonpres. - This is like having perfectly collinear variables and we cannot estimate both.
- This is one possible source of errors saying singular somewhere in the error.
Let’s try again, being a bit more conservative in the variables which are included:
bigReg <- lm(vitdresul ~ gend + age + vitdc + vit.12 +
vitbc + folate + TEON,
data=meddata, na.action = na.fail)
summary(bigReg)
Call:
lm(formula = vitdresul ~ gend + age + vitdc + vit.12 + vitbc +
folate + TEON, data = meddata, na.action = na.fail)
Residuals:
Min 1Q Median 3Q Max
-6.604 -2.603 0.000 1.869 10.845
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 8.415038 7.653793 1.099 0.276940
gendmale 0.561497 1.269939 0.442 0.660330
age -0.011608 0.042999 -0.270 0.788315
vitdcinsufficiency 7.186854 1.814761 3.960 0.000242 ***
vitdclow 6.451302 4.585442 1.407 0.165766
vitdcnormal 23.705316 3.405476 6.961 7.63e-09 ***
vit.12 0.000514 0.005760 0.089 0.929249
vitbclow 1.036309 8.309493 0.125 0.901260
vitbcnormal 2.811365 5.600409 0.502 0.617921
folate 0.060777 0.078379 0.775 0.441811
TEONYes -5.155763 1.494984 -3.449 0.001167 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.963 on 49 degrees of freedom
Multiple R-squared: 0.7162, Adjusted R-squared: 0.6583
F-statistic: 12.37 on 10 and 49 DF, p-value: 2.451e-1015.7.4 What terms are significant?
We can look to get overall tests for the components (rather than examining the individual factor-level estimates).
anova(bigReg)Analysis of Variance Table
Response: vitdresul
Df Sum Sq Mean Sq F value Pr(>F)
gend 1 157.32 157.32 10.0155 0.002668 **
age 1 38.42 38.42 2.4459 0.124270
vitdc 3 1515.01 505.00 32.1493 1.236e-11 ***
vit.12 1 29.15 29.15 1.8556 0.179358
vitbc 2 11.43 5.72 0.3639 0.696845
folate 1 4.45 4.45 0.2834 0.596906
TEON 1 186.83 186.83 11.8936 0.001167 **
Residuals 49 769.70 15.71
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- As we have seen previously, with
anovathe order of the variables is important (technically we’d have to work from the bottom up in interpretation). - A more practical version is the
Anovacommand in thecarlibrary as mentioned previously.
Anova(bigReg)Anova Table (Type II tests)
Response: vitdresul
Sum Sq Df F value Pr(>F)
gend 3.07 1 0.1955 0.660330
age 1.14 1 0.0729 0.788315
vitdc 764.87 3 16.2310 1.857e-07 ***
vit.12 0.13 1 0.0080 0.929249
vitbc 5.77 2 0.1836 0.832837
folate 9.45 1 0.6013 0.441811
TEON 186.83 1 11.8936 0.001167 **
Residuals 769.70 49
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1So what to keep?
- Going backwards, we might drop the least significant - here
vit.12, by the ‘-vit.12’ indicating removevit.12variable. - We can use the
updatefunction to drop/add terms
smallerReg <- update(bigReg, .~.-vit.12)
Anova(smallerReg)Anova Table (Type II tests)
Response: vitdresul
Sum Sq Df F value Pr(>F)
gend 2.96 1 0.1924 0.6628101
age 1.15 1 0.0747 0.7858029
vitdc 769.37 3 16.6569 1.238e-07 ***
vitbc 7.06 2 0.2293 0.7959354
folate 10.81 1 0.7023 0.4060049
TEON 197.87 1 12.8518 0.0007642 ***
Residuals 769.82 50
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1So what to keep?
- Applying the same rationale - drop
vitbc
smallerReg <- update(smallerReg, .~.-vitbc)
Anova(smallerReg)So on and so forth …
Anova Table (Type II tests)
Response: vitdresul
Sum Sq Df F value Pr(>F)
gend 1.86 1 0.1247 0.725381
age 1.08 1 0.0722 0.789277
vitdc 763.67 3 17.0384 7.787e-08 ***
folate 12.12 1 0.8115 0.371830
TEON 223.66 1 14.9702 0.000306 ***
Residuals 776.88 52
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Applying the same rationale - we now drop
age
evensmallerReg <- update(smallerReg, .~.-age)
Anova(evensmallerReg)Anova Table (Type II tests)
Response: vitdresul
Sum Sq Df F value Pr(>F)
gend 1.97 1 0.1340 0.7157421
vitdc 808.05 3 18.3499 2.713e-08 ***
folate 13.25 1 0.9028 0.3463528
TEON 224.41 1 15.2881 0.0002641 ***
Residuals 777.96 53
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Applying the same rationale again - we now drop
gend
evenevensmallerReg <- update(evensmallerReg, .~.-gend)
Anova(evenevensmallerReg)Anova Table (Type II tests)
Response: vitdresul
Sum Sq Df F value Pr(>F)
vitdc 806.20 3 18.6062 2.046e-08 ***
folate 11.59 1 0.8027 0.3742660
TEON 226.59 1 15.6885 0.0002204 ***
Residuals 779.93 54
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Applying the same rationale again - we now drop
folate
evenevenevensmallerReg <- update(evenevensmallerReg, .~.-folate)
Anova(evenevenevensmallerReg)Anova Table (Type II tests)
Response: vitdresul
Sum Sq Df F value Pr(>F)
vitdc 842.24 3 19.508 9.686e-09 ***
TEON 216.55 1 15.047 0.0002825 ***
Residuals 791.52 55
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1All the terms in this model are associated with a \(p\)-value <0.05 and so are retained.
15.7.5 More automated methods
Rather than fitting models, examining output, changing the model and refitting etc. procedures exist that do all this automatically.
15.7.5.1 Stepwise selection by AIC
Rather than using the \(p\)-value for model selection, we saw other criteria such as AIC might be used. We can use the step function to apply these automatically:
smallerReg <- step(bigReg)Start: AIC=175.1
vitdresul ~ gend + age + vitdc + vit.12 + vitbc + folate + TEON
Df Sum of Sq RSS AIC
- vitbc 2 5.77 775.47 171.55
- vit.12 1 0.13 769.82 173.11
- age 1 1.14 770.84 173.19
- gend 1 3.07 772.77 173.34
- folate 1 9.45 779.14 173.83
<none> 769.70 175.10
- TEON 1 186.83 956.52 186.14
- vitdc 3 764.87 1534.57 210.50
Step: AIC=171.55
vitdresul ~ gend + age + vitdc + vit.12 + folate + TEON
Df Sum of Sq RSS AIC
- age 1 1.17 776.64 169.64
- vit.12 1 1.42 776.88 169.66
- gend 1 1.97 777.43 169.70
- folate 1 13.50 788.97 170.58
<none> 775.47 171.55
- TEON 1 188.92 964.39 182.63
- vitdc 3 763.04 1538.50 206.65
Step: AIC=169.64
vitdresul ~ gend + vitdc + vit.12 + folate + TEON
Df Sum of Sq RSS AIC
- vit.12 1 1.32 777.96 167.74
- gend 1 2.07 778.71 167.80
- folate 1 14.57 791.20 168.75
<none> 776.64 169.64
- TEON 1 188.42 965.06 180.67
- vitdc 3 806.87 1583.50 206.38
Step: AIC=167.74
vitdresul ~ gend + vitdc + folate + TEON
Df Sum of Sq RSS AIC
- gend 1 1.97 779.93 165.89
- folate 1 13.25 791.21 166.75
<none> 777.96 167.74
- TEON 1 224.41 1002.37 180.95
- vitdc 3 808.05 1586.01 204.48
Step: AIC=165.89
vitdresul ~ vitdc + folate + TEON
Df Sum of Sq RSS AIC
- folate 1 11.59 791.52 164.78
<none> 779.93 165.89
- TEON 1 226.59 1006.52 179.19
- vitdc 3 806.20 1586.12 202.48
Step: AIC=164.78
vitdresul ~ vitdc + TEON
Df Sum of Sq RSS AIC
<none> 791.52 164.78
- TEON 1 216.55 1008.07 177.29
- vitdc 3 842.24 1633.76 202.26summary(smallerReg)
Call:
lm(formula = vitdresul ~ vitdc + TEON, data = meddata, na.action = na.fail)
Residuals:
Min 1Q Median 3Q Max
-6.9430 -2.5992 -0.2422 1.9645 10.5070
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 11.583 1.666 6.954 4.49e-09 ***
vitdcinsufficiency 7.350 1.588 4.629 2.28e-05 ***
vitdclow 6.253 2.941 2.126 0.038009 *
vitdcnormal 23.762 3.158 7.525 5.20e-10 ***
TEONYes -4.982 1.284 -3.879 0.000282 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.794 on 55 degrees of freedom
Multiple R-squared: 0.7082, Adjusted R-squared: 0.687
F-statistic: 33.37 on 4 and 55 DF, p-value: 4e-14Anova(smallerReg)Anova Table (Type II tests)
Response: vitdresul
Sum Sq Df F value Pr(>F)
vitdc 842.24 3 19.508 9.686e-09 ***
TEON 216.55 1 15.047 0.0002825 ***
Residuals 791.52 55
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- We can see at each step all the covariates are considered for exclusion
- The exclusion that gives the lowest AIC is favoured
- The process repeats on this reduced model
- The process terminates when deletions no longer improve the AIC
- We don’t necessarily get the same model as other methods e.g. \(p\)-value deletion. AIC is a different model selection criterion.
15.7.5.2 All possible subsets
The previous method was an incomplete search - not all models considered. The best model at any point, depends on the previous step, and although efficient is not guaranteed to find our best collection of covariates within our big model specified at the start.
We can try all possible models (combining up to say 60 terms).
- Remember the
dredgecommand for this in theMuMInpackage:
library(MuMIn)
dredgedReg<- dredge(bigReg)
dredgedRegGlobal model call: lm(formula = vitdresul ~ gend + age + vitdc + vit.12 + vitbc +
folate + TEON, data = meddata, na.action = na.fail)
---
Model selection table
(Intr) age folt gend TEON vit.12 vtbc vtdc df logLik AICc delta weight
73 11.5800 + + 6 -162.525 338.6 0.00 0.305
75 11.1900 0.06068 + + 7 -162.082 340.3 1.68 0.131
74 12.3700 -0.015980 + + 7 -162.441 341.0 2.40 0.092
77 11.5300 + + + 7 -162.513 341.2 2.55 0.085
89 11.6100 + -1.121e-04 + 7 -162.524 341.2 2.57 0.084
79 11.0100 0.06677 + + + 8 -162.006 342.8 4.20 0.037
91 11.4700 0.06642 + -1.215e-03 + 8 -162.035 342.9 4.26 0.036
76 11.7800 -0.011760 0.05834 + + 8 -162.037 342.9 4.26 0.036
105 9.5110 + + + 8 -162.218 343.3 4.63 0.030
78 12.3200 -0.015940 + + + 8 -162.429 343.7 5.05 0.024
90 12.4400 -0.016140 + -2.239e-04 + 8 -162.439 343.7 5.07 0.024
93 11.5600 + + -9.141e-05 + 8 -162.513 343.8 5.21 0.022
107 8.9870 0.05632 + + + 9 -161.854 345.3 6.67 0.011
95 11.3100 0.07292 + + -1.268e-03 + 9 -161.955 345.5 6.88 0.010
80 11.5900 -0.011230 0.06437 + + + 9 -161.965 345.5 6.90 0.010
92 12.1000 -0.012230 0.06421 + -1.263e-03 + 9 -161.986 345.6 6.94 0.009
106 10.1800 -0.015300 + + + 9 -162.141 345.9 7.25 0.008
121 8.1360 + 1.464e-03 + + 9 -162.174 345.9 7.31 0.008
109 9.2390 + + + + 9 -162.187 346.0 7.34 0.008
94 12.3800 -0.016090 + + -2.033e-04 + 9 -162.428 346.5 7.82 0.006
111 8.3630 0.06478 + + + + 10 -161.736 348.0 9.33 0.003
108 9.5290 -0.012040 0.05424 + + + 10 -161.806 348.1 9.47 0.003
123 8.8150 0.05554 + 1.910e-04 + + 10 -161.853 348.2 9.56 0.003
96 11.9200 -0.011720 0.07064 + + -1.313e-03 + 10 -161.910 348.3 9.68 0.002
122 8.8450 -0.014930 + 1.399e-03 + + 10 -162.101 348.7 10.06 0.002
110 9.9030 -0.015350 + + + + 10 -162.109 348.7 10.07 0.002
125 7.4940 + + 1.776e-03 + + 10 -162.125 348.7 10.11 0.002
65 7.1000 + 5 -169.780 350.7 12.04 0.001
112 8.8960 -0.011630 0.06265 + + + + 11 -161.691 350.9 12.25 0.001
127 7.8750 0.06287 + + 5.232e-04 + + 11 -161.730 351.0 12.33 0.001
124 9.3610 -0.012040 0.05348 + 1.862e-04 + + 11 -161.805 351.1 12.48 0.001
81 9.1860 -5.486e-03 + 6 -168.834 351.3 12.62 0.001
126 8.2030 -0.014900 + + 1.711e-03 + + 11 -162.052 351.6 12.97 0.000
69 7.4330 + + 6 -169.629 352.8 14.21 0.000
66 6.4960 0.013720 + 6 -169.729 353.0 14.41 0.000
67 6.9020 0.02198 + 6 -169.734 353.1 14.42 0.000
83 9.0430 0.05829 -6.494e-03 + 7 -168.529 353.2 14.58 0.000
97 1.3350 + + 7 -168.651 353.5 14.82 0.000
85 9.4590 + -5.409e-03 + 7 -168.706 353.6 14.93 0.000
82 8.8980 0.005911 -5.412e-03 + 7 -168.824 353.8 15.17 0.000
128 8.4150 -0.011610 0.06078 + + 5.140e-04 + + 12 -161.686 354.0 15.38 0.000
70 6.8640 0.012770 + + 7 -169.585 355.3 16.69 0.000
71 7.2770 0.01454 + + 7 -169.610 355.4 16.74 0.000
68 6.1660 0.015940 0.02581 + 7 -169.667 355.5 16.85 0.000
113 5.1540 -3.592e-03 + + 8 -168.421 355.7 17.03 0.000
87 9.2450 0.05244 + -6.340e-03 + 8 -168.472 355.8 17.13 0.000
84 8.5690 0.009647 0.06011 -6.405e-03 + 8 -168.504 355.8 17.20 0.000
99 1.0250 0.02272 + + 8 -168.603 356.0 17.39 0.000
101 1.7200 + + + 8 -168.619 356.1 17.43 0.000
98 1.0110 0.009511 + + 8 -168.626 356.1 17.44 0.000
86 9.2060 0.005134 + -5.345e-03 + 8 -168.699 356.2 17.59 0.000
72 6.5920 0.014440 0.01837 + + 8 -169.555 357.9 19.30 0.000
115 5.6640 0.04361 -4.630e-03 + + 9 -168.260 358.1 19.49 0.000
117 6.0100 + -3.877e-03 + + 9 -168.357 358.3 19.68 0.000
114 4.8310 0.007293 -3.521e-03 + + 9 -168.407 358.4 19.78 0.000
88 8.8050 0.008769 0.05432 + -6.265e-03 + 9 -168.451 358.5 19.87 0.000
100 0.6089 0.011300 0.02507 + + 9 -168.568 358.7 20.10 0.000
103 1.3590 0.01951 + + + 9 -168.586 358.8 20.14 0.000
102 1.3970 0.009314 + + + 9 -168.596 358.8 20.16 0.000
119 6.2320 0.03973 + -4.742e-03 + + 10 -168.229 360.9 22.31 0.000
116 5.2480 0.009862 0.04542 -4.578e-03 + + 10 -168.234 361.0 22.32 0.000
118 5.6960 0.006830 + -3.807e-03 + + 10 -168.344 361.2 22.54 0.000
104 0.9333 0.010940 0.02200 + + + 10 -168.553 361.6 22.96 0.000
120 5.8130 0.009326 0.04162 + -4.688e-03 + + 11 -168.205 363.9 25.28 0.000
9 19.5900 + 3 -184.265 375.0 36.32 0.000
10 22.5100 -0.078460 + 4 -183.121 375.0 36.33 0.000
11 18.2400 0.12040 + 4 -183.377 375.5 36.85 0.000
12 21.0300 -0.068340 0.09939 + 5 -182.515 376.1 37.51 0.000
13 19.7000 + + 4 -184.198 377.1 38.49 0.000
14 22.6600 -0.078980 + + 5 -183.036 377.2 38.55 0.000
25 19.5000 + 2.853e-04 4 -184.264 377.3 38.62 0.000
26 22.4800 -0.078440 + 1.128e-04 5 -183.121 377.4 38.72 0.000
27 18.6300 0.12780 + -1.628e-03 5 -183.331 377.8 39.14 0.000
15 18.2800 0.11900 + + 5 -183.375 377.9 39.23 0.000
28 21.3600 -0.067980 0.10610 + -1.453e-03 6 -182.477 378.5 39.91 0.000
16 21.1300 -0.068900 0.09592 + + 6 -182.503 378.6 39.96 0.000
29 19.6700 + + 9.175e-05 5 -184.197 379.5 40.87 0.000
41 21.2100 + + 5 -184.212 379.5 40.90 0.000
30 22.6900 -0.079000 + + -1.033e-04 6 -183.036 379.7 41.02 0.000
42 23.3000 -0.077620 + + 6 -183.109 379.8 41.17 0.000
43 19.3200 0.12270 + + 6 -183.309 380.2 41.57 0.000
31 18.6800 0.12620 + + -1.647e-03 6 -183.328 380.2 41.61 0.000
32 21.4800 -0.068570 0.10250 + + -1.491e-03 7 -182.463 381.1 42.45 0.000
44 21.4500 -0.066970 0.10130 + + 7 -182.496 381.1 42.51 0.000
45 21.7200 + + + 6 -184.114 381.8 43.18 0.000
57 22.0300 + -8.985e-04 + 6 -184.203 382.0 43.36 0.000
46 23.8000 -0.077570 + + + 7 -183.010 382.2 43.54 0.000
58 23.8800 -0.077500 + -6.457e-04 + 7 -183.105 382.4 43.73 0.000
59 22.2200 0.13340 + -3.377e-03 + 7 -183.197 382.5 43.91 0.000
47 19.5300 0.11970 + + + 7 -183.299 382.8 44.12 0.000
60 23.7500 -0.065500 0.11040 + -2.736e-03 + 8 -182.421 383.7 45.03 0.000
48 21.7500 -0.067400 0.09698 + + + 8 -182.476 383.8 45.14 0.000
61 23.2400 + + -1.615e-03 + 7 -184.089 384.3 45.70 0.000
62 25.0600 -0.077310 + + -1.346e-03 + 8 -182.991 384.8 46.17 0.000
63 22.7700 0.12950 + + -3.636e-03 + 8 -183.173 385.2 46.53 0.000
64 24.4100 -0.065920 0.10570 + + -3.035e-03 + 9 -182.386 386.4 47.74 0.000
21 20.7700 + -9.925e-03 4 -196.362 401.5 62.82 0.000
5 17.5800 + 3 -197.680 401.8 63.15 0.000
22 23.3200 -0.066640 + -1.019e-02 5 -195.816 402.7 64.11 0.000
17 19.7500 -1.015e-02 3 -198.174 402.8 64.14 0.000
23 19.9300 0.10710 + -1.147e-02 5 -195.947 403.0 64.37 0.000
1 16.4700 2 -199.473 403.2 64.52 0.000
6 19.8800 -0.062120 + 4 -197.225 403.2 64.54 0.000
19 18.7300 0.14950 -1.228e-02 4 -197.371 403.5 64.84 0.000
7 16.9700 0.04860 + 4 -197.592 403.9 65.28 0.000
18 22.0800 -0.061120 -1.040e-02 4 -197.741 404.2 65.57 0.000
53 32.0400 + -1.652e-02 + 6 -195.456 404.5 65.86 0.000
2 18.5400 -0.056420 3 -199.119 404.7 66.03 0.000
24 22.3000 -0.057730 0.08705 + -1.141e-02 6 -195.547 404.7 66.04 0.000
3 15.4400 0.09022 3 -199.175 404.8 66.14 0.000
20 20.6600 -0.048080 0.13430 -1.226e-02 5 -197.105 405.3 66.69 0.000
8 19.4100 -0.059040 0.02842 + 5 -197.196 405.5 66.87 0.000
49 27.1200 -1.512e-02 + 5 -197.294 405.7 67.06 0.000
54 33.6900 -0.066420 + -1.642e-02 + 7 -194.906 406.0 67.33 0.000
55 31.7100 0.11170 + -1.837e-02 + 7 -194.992 406.1 67.50 0.000
37 15.4200 + + 5 -197.532 406.2 67.54 0.000
51 27.2900 0.14940 -1.778e-02 + 6 -196.480 406.5 67.91 0.000
4 17.4000 -0.048670 0.07498 4 -198.918 406.6 67.93 0.000
33 12.2100 + 4 -198.956 406.6 68.01 0.000
50 28.7400 -0.065720 -1.503e-02 + 6 -196.787 407.2 68.53 0.000
38 17.1900 -0.067870 + + 6 -196.996 407.6 68.94 0.000
34 13.9500 -0.067110 + 5 -198.456 408.0 69.39 0.000
56 33.1700 -0.056540 0.09117 + -1.795e-02 + 8 -194.604 408.0 69.40 0.000
35 10.7700 0.08768 + 5 -198.676 408.5 69.83 0.000
39 14.4300 0.05008 + + 6 -197.441 408.5 69.83 0.000
52 28.5400 -0.051550 0.13140 -1.739e-02 + 7 -196.173 408.5 69.87 0.000
40 16.5600 -0.064860 0.02813 + + 7 -196.968 410.1 71.46 0.000
36 12.6400 -0.059850 0.06840 + 6 -198.289 410.2 71.53 0.000
Models ranked by AICc(x) This produces a lot of output. We see:
- The models are ranked on the basis of AICc
- We have coefficients for some covariates, not others - these are the excluded covariates
- We have a \(\Delta\) AICc which shows how much the AICc has changed
- We could choose the best model and use it or
- [For a predictive model, we could average a number of these, weighted by AIC - but that is for another course.]
Let’s only consider the ‘best’ models from the set of all possible models, i.e. select the best model and all models with a \(\Delta\) AICc < 5 of the best model.
topMods <- get.models(dredgedReg, subset=delta<5)
summary(topMods[[1]])
Call:
lm(formula = vitdresul ~ TEON + vitdc + 1, data = meddata, na.action = na.fail)
Residuals:
Min 1Q Median 3Q Max
-6.9430 -2.5992 -0.2422 1.9645 10.5070
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 11.583 1.666 6.954 4.49e-09 ***
TEONYes -4.982 1.284 -3.879 0.000282 ***
vitdcinsufficiency 7.350 1.588 4.629 2.28e-05 ***
vitdclow 6.253 2.941 2.126 0.038009 *
vitdcnormal 23.762 3.158 7.525 5.20e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.794 on 55 degrees of freedom
Multiple R-squared: 0.7082, Adjusted R-squared: 0.687
F-statistic: 33.37 on 4 and 55 DF, p-value: 4e-14The object topMods is a list of models. Items in a list are specified by two square brackets. The best model is in position 1.
The use of +1 in the model formula here refers to a normally unstated default in R. +1 one just means an intercept should be calculated.
Q15.2 Dr X is investigating whether human height can be predicted from measured leg length (again!) and, in addition, index finger length. The data collected are as follows: left leg length (LLL), right leg length (RLL), right finger length (finger), total height (height) and sex at birth (Sex). All length measurements are in cm. Dr X fits the following model.
Model3 <- lm(height ~ LLL + RLL + finger + Sex, data=df1)Write down the general equation for this model.
Q15.3 The summary of the model fitted in Q15.2 is shown below. State the null and alternative hypotheses for testing each regression coefficient.
Call:
lm(formula = height ~ LLL + RLL + finger + Sex)
Residuals:
Min 1Q Median 3Q Max
-4.2560 -0.9569 -0.1777 1.2501 4.7053
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.54854 4.69342 -0.543 0.588
LLL 0.49232 0.35872 1.372 0.173
RLL 0.47476 0.36618 1.297 0.198
finger 0.08138 0.17294 0.471 0.639
Sex -0.81689 0.61429 -1.330 0.187
Residual standard error: 1.743 on 95 degrees of freedom
Multiple R-squared: 0.877, Adjusted R-squared: 0.8718
F-statistic: 169.3 on 4 and 95 DF, p-value: < 2.2e-1615.4 Are any of the explanatory variables significant, testing at a 5% fixed significance level, based on the t-statistics?
15.5 The VIF analysis associated with the model is below.
vif(model1)
LLL RLL finger Sex
94.606522 95.274197 1.077881 1.016933What do you conclude from this VIF analysis and model summary above?
Q15.6 How could you further investigate relationships between explanatory variables?
Q15.7 Independently of Dr X, Prof Y is also analysing the data. Prof Y’s modelling philosophy is to use AIC for model selection and obtains the following information.
model1 <- lm (height ~ LLL + RLL + finger + Sex)
> AIC (model1)
[1] 401.5684
> model2a <- lm (height ~ RLL + finger + Sex)
> AIC (model2a)
[1] 401.7778
> model2b <- lm (height ~ LLL + finger + Sex)
> AIC (model2b)
[1] 401.5684
> model2c <- lm (height ~ LLL + RLL + Sex)
> AIC (model2c)
[1] 400.0473
> model2d <- lm (height ~ LLL + RLL + finger)
> AIC (model2d)
[1] 401.6588Using this information, what would you do next in the modelling process?
15.8 Summary
A final note on model selection:
- Be very cautious of automatic model selection tools if you intend to describe/interpret your model because the retention of the variables in an automatic process is really directed towards prediction (rather than explanation) of the observed data.
- Small perturbations in the data (or indeed another sample) can produce a wildly different model structure although the predictions themselves might be quite similar.
Note that implicit in this chapter, is that we want a model for prediction or understanding. In contrast, it may be the whole purpose of the model is to test a particular hypothesis. In this case, model reduction may not be necessary. What needs to be obtained is the probability associated with particular variable of interest as its significance is what is reported. However, we still may want to eliminate extraneous variables to make that particular test as efficient as possible.
15.8.1 Learning objectives
At the end of this chapter you should understand:
- why model selection can be important, and
- how model selection is undertaken.
15.9 Answers
Q15.1 A low VIF does not indicate a variable should be in the model, merely that it is not correlated with another predictor. Its inclusion in the model should still be tested.
Q15.2 \[ height=\ {\beta{}}_0+\ {\beta{}}_1LLL+\ {\beta{}}_2RLL+\ {\beta{}}_3finger+\ {\beta{}}_4Sex+\ \epsilon{} \]
Q15.3 The hypotheses are: \(H_{0}\): \({\beta{}}_p=0\) (i.e. no relationship between explanatory variable \(p\) and the response variable, height) and
\(H_{1}\): \({\beta{}}_p\not=0\) (i.e. there is a relationship between variable \(p\) and the response and hence, variable is a useful explanatory variable)
Q15.4 No! All probabilities (Pr(>|t|)) are greater than 0.05.
Q15.5 The VIF scores (and common sense) imply that LLL and RLL are collinear and this correlation leads to a failure to detect a significant leg length effect. Therefore, one of the leg lengths should be removed from the model.
Q15.6 Plotting the explanatory variables against each other would indicate the relationships between them. The pairs function is a useful function for doing this.
Q15.7 The better fitting models are the ones with the lowest AIC. Therefore, reject finger as an explanatory variable because the model without finger (i.e. model2c) has the lowest AIC; refit the models using the reduced set of explanatory variables, check the AIC values for the new models.